Simplifying lists
library(tidyverse)
library(httr)
library(repurrrsive)
set.seed(123)
theme_set(theme_minimal())
Run the code below in your console to download this exercise as a set of R scripts.
usethis::use_course("cis-ds/getting-data-from-the-web-api-access")
Not all lists are easily coerced into data frames by simply calling content() %>% as_tibble(). Unless your list is perfectly structured, this will not work. Recall the OMDB example:
sharknado <- GET(
url = "http://www.omdbapi.com/?",
query = list(
t = "Sharknado",
y = 2013,
apikey = getOption("omdb_key")
)
)
# convert to data frame
content(sharknado, as = "parsed", type = "application/json") %>%
as_tibble()
## # A tibble: 2 x 25
## Title Year Rated Released Runtime Genre Director Writer Actors Plot Language
## <chr> <chr> <chr> <chr> <chr> <chr> <chr> <chr> <chr> <chr> <chr>
## 1 Shar… 2013 TV-14 11 Jul … 86 min Acti… Anthony… Thund… Ian Z… When… English
## 2 Shar… 2013 TV-14 11 Jul … 86 min Acti… Anthony… Thund… Ian Z… When… English
## # … with 14 more variables: Country <chr>, Awards <chr>, Poster <chr>,
## # Ratings <list>, Metascore <chr>, imdbRating <chr>, imdbVotes <chr>,
## # imdbID <chr>, Type <chr>, DVD <chr>, BoxOffice <chr>, Production <chr>,
## # Website <chr>, Response <chr>
Wait a minute, what happened? Look at the structure of content():
content(sharknado) %>%
str()
## List of 25
## $ Title : chr "Sharknado"
## $ Year : chr "2013"
## $ Rated : chr "TV-14"
## $ Released : chr "11 Jul 2013"
## $ Runtime : chr "86 min"
## $ Genre : chr "Action, Adventure, Comedy, Horror, Sci-Fi, Thriller"
## $ Director : chr "Anthony C. Ferrante"
## $ Writer : chr "Thunder Levin"
## $ Actors : chr "Ian Ziering, Tara Reid, John Heard, Cassandra Scerbo"
## $ Plot : chr "When a freak hurricane swamps Los Angeles, nature's deadliest killer rules sea, land, and air as thousands of s"| __truncated__
## $ Language : chr "English"
## $ Country : chr "USA"
## $ Awards : chr "1 win & 2 nominations."
## $ Poster : chr "https://m.media-amazon.com/images/M/MV5BODcwZWFiNTEtNDgzMC00ZmE2LWExMzYtNzZhZDgzNDc5NDkyXkEyXkFqcGdeQXVyMTQxNzM"| __truncated__
## $ Ratings :List of 2
## ..$ :List of 2
## .. ..$ Source: chr "Internet Movie Database"
## .. ..$ Value : chr "3.3/10"
## ..$ :List of 2
## .. ..$ Source: chr "Rotten Tomatoes"
## .. ..$ Value : chr "74%"
## $ Metascore : chr "N/A"
## $ imdbRating: chr "3.3"
## $ imdbVotes : chr "46,752"
## $ imdbID : chr "tt2724064"
## $ Type : chr "movie"
## $ DVD : chr "11 Oct 2016"
## $ BoxOffice : chr "N/A"
## $ Production: chr "The Asylum, Southward Films"
## $ Website : chr "N/A"
## $ Response : chr "True"
Look at the ratings element: it is a data frame. Remember that data frames are just a special type of list, so what we have here is a list inside of a list (aka a recursive list). We cannot easily flatten this into a data frame, because the ratings element is not an atomic vector of length 1 like all the other elements in sharknado. Instead, we have to think of another way to convert it to a data frame.
Rectangling and tidyr
Rectangling is the art and craft of taking a deeply nested list (often sourced from wild caught JSON or XML) and taming it into a tidy data set of rows and columns. There are three functions from tidyr that are particularly useful for rectangling:
unnest_longer()takes each element of a list-column and makes a new row.unnest_wider()takes each element of a list-column and makes a new column.unnest_auto()guesses whether you wantunnest_longer()orunnest_wider().hoist()is similar tounnest_wider()but only plucks out selected components, and can reach down multiple levels.
A very large number of data rectangling problems can be solved by combining these functions with a splash of dplyr.
Load packages
We need to load two packages now: repurrrsive contains examples of recursive lists, and listviewer which provides an interactive method for viewing the structure of a list.
remotes::install_github("jennybc/repurrrsive")
install.packages("listviewer")
library(purrr)
library(repurrrsive)
Inspecting and exploring lists
Before you can apply functions to a list, you should understand it. Especially when dealing with poorly documented APIs, you may not know in advance the structure of your list, or it may not be the same as the documentation. str() is the base R method for inspecting a list by printing the structure of the list to the console. If you have a large list, this will be a lot of output. max.levels and list.len can be used to print only a partial structure for this list.
listviewer::jsonedit() to interactively view the list within RStudio.unnest_wider() and hoist()
Let’s look at gh_users which is a list that contains information about six GitHub users.
str(gh_users, list.len = 3)
## List of 6
## $ :List of 30
## ..$ login : chr "gaborcsardi"
## ..$ id : int 660288
## ..$ avatar_url : chr "https://avatars.githubusercontent.com/u/660288?v=3"
## .. [list output truncated]
## $ :List of 30
## ..$ login : chr "jennybc"
## ..$ id : int 599454
## ..$ avatar_url : chr "https://avatars.githubusercontent.com/u/599454?v=3"
## .. [list output truncated]
## $ :List of 30
## ..$ login : chr "jtleek"
## ..$ id : int 1571674
## ..$ avatar_url : chr "https://avatars.githubusercontent.com/u/1571674?v=3"
## .. [list output truncated]
## [list output truncated]
To begin, we first put gh_users into a data frame:
(users <- tibble(user = gh_users))
## # A tibble: 6 x 1
## user
## <list>
## 1 <named list [30]>
## 2 <named list [30]>
## 3 <named list [30]>
## 4 <named list [30]>
## 5 <named list [30]>
## 6 <named list [30]>
We’ve already seen examples of list-columns. By storing the list in a data frame, we bundle together multiple vectors so when we start to extract elements they are stored in a single object.
Each user is a named list, where each element represents a column:
names(users$user[[1]])
## [1] "login" "id" "avatar_url"
## [4] "gravatar_id" "url" "html_url"
## [7] "followers_url" "following_url" "gists_url"
## [10] "starred_url" "subscriptions_url" "organizations_url"
## [13] "repos_url" "events_url" "received_events_url"
## [16] "type" "site_admin" "name"
## [19] "company" "blog" "location"
## [22] "email" "hireable" "bio"
## [25] "public_repos" "public_gists" "followers"
## [28] "following" "created_at" "updated_at"
There are two ways to turn the list components into columns. unnest_wider() takes every component and makes a new column:
users %>%
unnest_wider(user)
## # A tibble: 6 x 30
## login id avatar_url gravatar_id url html_url followers_url following_url
## <chr> <int> <chr> <chr> <chr> <chr> <chr> <chr>
## 1 gabo… 6.60e5 https://a… "" http… https:/… https://api.… https://api.…
## 2 jenn… 5.99e5 https://a… "" http… https:/… https://api.… https://api.…
## 3 jtle… 1.57e6 https://a… "" http… https:/… https://api.… https://api.…
## 4 juli… 1.25e7 https://a… "" http… https:/… https://api.… https://api.…
## 5 leep… 3.51e6 https://a… "" http… https:/… https://api.… https://api.…
## 6 masa… 8.36e6 https://a… "" http… https:/… https://api.… https://api.…
## # … with 22 more variables: gists_url <chr>, starred_url <chr>,
## # subscriptions_url <chr>, organizations_url <chr>, repos_url <chr>,
## # events_url <chr>, received_events_url <chr>, type <chr>, site_admin <lgl>,
## # name <chr>, company <chr>, blog <chr>, location <chr>, email <chr>,
## # public_repos <int>, public_gists <int>, followers <int>, following <int>,
## # created_at <chr>, updated_at <chr>, bio <chr>, hireable <lgl>
Extremely easy! However, there are a lot of components in users, and we don’t necessarily want or need all of them. Instead, we can use hoist() to pull out selected components:
users %>%
hoist(user,
followers = "followers",
login = "login",
url = "html_url"
)
## # A tibble: 6 x 4
## followers login url user
## <int> <chr> <chr> <list>
## 1 303 gaborcsardi https://github.com/gaborcsardi <named list [27]>
## 2 780 jennybc https://github.com/jennybc <named list [27]>
## 3 3958 jtleek https://github.com/jtleek <named list [27]>
## 4 115 juliasilge https://github.com/juliasilge <named list [27]>
## 5 213 leeper https://github.com/leeper <named list [27]>
## 6 34 masalmon https://github.com/masalmon <named list [27]>
hoist() removes the named components from the user list-column while retaining the unnamed components, so it’s equivalent to moving the components out of the inner list into the top-level data frame.
gh_repos and nested list structures
We start off gh_repos similarly, by putting it in a tibble:
(repos <- tibble(repo = gh_repos))
## # A tibble: 6 x 1
## repo
## <list>
## 1 <list [30]>
## 2 <list [30]>
## 3 <list [30]>
## 4 <list [26]>
## 5 <list [30]>
## 6 <list [30]>
This time the elements of repo are a list of repositories that belong to that user. These are observations, so should become new rows, so we use unnest_longer() rather than unnest_wider():
repos <- repos %>%
unnest_longer(repo)
repos
## # A tibble: 176 x 1
## repo
## <list>
## 1 <named list [68]>
## 2 <named list [68]>
## 3 <named list [68]>
## 4 <named list [68]>
## 5 <named list [68]>
## 6 <named list [68]>
## 7 <named list [68]>
## 8 <named list [68]>
## 9 <named list [68]>
## 10 <named list [68]>
## # … with 166 more rows
Then we can use unnest_wider() or hoist():
repos %>%
hoist(repo,
login = c("owner", "login"),
name = "name",
homepage = "homepage",
watchers = "watchers_count"
)
## # A tibble: 176 x 5
## login name homepage watchers repo
## <chr> <chr> <chr> <int> <list>
## 1 gaborcsardi after <NA> 5 <named list [65]>
## 2 gaborcsardi argufy <NA> 19 <named list [65]>
## 3 gaborcsardi ask <NA> 5 <named list [65]>
## 4 gaborcsardi baseimports <NA> 0 <named list [65]>
## 5 gaborcsardi citest <NA> 0 <named list [65]>
## 6 gaborcsardi clisymbols "" 18 <named list [65]>
## 7 gaborcsardi cmaker <NA> 0 <named list [65]>
## 8 gaborcsardi cmark <NA> 0 <named list [65]>
## 9 gaborcsardi conditions <NA> 0 <named list [65]>
## 10 gaborcsardi crayon <NA> 52 <named list [65]>
## # … with 166 more rows
Note the use of c("owner", "login"): this allows us to reach two levels deep inside of a list. An alternative approach would be to pull out just owner and then put each element of it in a column:
repos %>%
hoist(repo, owner = "owner") %>%
unnest_wider(owner)
## # A tibble: 176 x 18
## login id avatar_url gravatar_id url html_url followers_url
## <chr> <int> <chr> <chr> <chr> <chr> <chr>
## 1 gabor… 660288 https://avata… "" https:/… https://g… https://api.git…
## 2 gabor… 660288 https://avata… "" https:/… https://g… https://api.git…
## 3 gabor… 660288 https://avata… "" https:/… https://g… https://api.git…
## 4 gabor… 660288 https://avata… "" https:/… https://g… https://api.git…
## 5 gabor… 660288 https://avata… "" https:/… https://g… https://api.git…
## 6 gabor… 660288 https://avata… "" https:/… https://g… https://api.git…
## 7 gabor… 660288 https://avata… "" https:/… https://g… https://api.git…
## 8 gabor… 660288 https://avata… "" https:/… https://g… https://api.git…
## 9 gabor… 660288 https://avata… "" https:/… https://g… https://api.git…
## 10 gabor… 660288 https://avata… "" https:/… https://g… https://api.git…
## # … with 166 more rows, and 11 more variables: following_url <chr>,
## # gists_url <chr>, starred_url <chr>, subscriptions_url <chr>,
## # organizations_url <chr>, repos_url <chr>, events_url <chr>,
## # received_events_url <chr>, type <chr>, site_admin <lgl>, repo <list>
Instead of looking at the list and carefully thinking about whether it needs to become rows or columns, you can use unnest_auto(). It uses a handful of heuristics to figure out whether unnest_longer() or unnest_wider() is appropriate, and tells you about its reasoning.
tibble(repo = gh_repos) %>%
unnest_auto(repo) %>%
unnest_auto(repo)
## Using `unnest_longer(repo)`; no element has names
## Using `unnest_wider(repo)`; elements have 68 names in common
## # A tibble: 176 x 67
## id name full_name owner private html_url description fork url
## <int> <chr> <chr> <lis> <lgl> <chr> <chr> <lgl> <chr>
## 1 6.12e7 after gaborcsar… <nam… FALSE https://… Run Code in … FALSE https:…
## 2 4.05e7 argufy gaborcsar… <nam… FALSE https://… Declarative … FALSE https:…
## 3 3.64e7 ask gaborcsar… <nam… FALSE https://… Friendly CLI… FALSE https:…
## 4 3.49e7 basei… gaborcsar… <nam… FALSE https://… Do we get wa… FALSE https:…
## 5 6.16e7 citest gaborcsar… <nam… FALSE https://… Test R packa… TRUE https:…
## 6 3.39e7 clisy… gaborcsar… <nam… FALSE https://… Unicode symb… FALSE https:…
## 7 3.72e7 cmaker gaborcsar… <nam… FALSE https://… port of cmak… TRUE https:…
## 8 6.80e7 cmark gaborcsar… <nam… FALSE https://… CommonMark p… TRUE https:…
## 9 6.32e7 condi… gaborcsar… <nam… FALSE https://… <NA> TRUE https:…
## 10 2.43e7 crayon gaborcsar… <nam… FALSE https://… R package fo… FALSE https:…
## # … with 166 more rows, and 58 more variables: forks_url <chr>, keys_url <chr>,
## # collaborators_url <chr>, teams_url <chr>, hooks_url <chr>,
## # issue_events_url <chr>, events_url <chr>, assignees_url <chr>,
## # branches_url <chr>, tags_url <chr>, blobs_url <chr>, git_tags_url <chr>,
## # git_refs_url <chr>, trees_url <chr>, statuses_url <chr>,
## # languages_url <chr>, stargazers_url <chr>, contributors_url <chr>,
## # subscribers_url <chr>, subscription_url <chr>, commits_url <chr>,
## # git_commits_url <chr>, comments_url <chr>, issue_comment_url <chr>,
## # contents_url <chr>, compare_url <chr>, merges_url <chr>, archive_url <chr>,
## # downloads_url <chr>, issues_url <chr>, pulls_url <chr>,
## # milestones_url <chr>, notifications_url <chr>, labels_url <chr>,
## # releases_url <chr>, deployments_url <chr>, created_at <chr>,
## # updated_at <chr>, pushed_at <chr>, git_url <chr>, ssh_url <chr>,
## # clone_url <chr>, svn_url <chr>, size <int>, stargazers_count <int>,
## # watchers_count <int>, language <chr>, has_issues <lgl>,
## # has_downloads <lgl>, has_wiki <lgl>, has_pages <lgl>, forks_count <int>,
## # open_issues_count <int>, forks <int>, open_issues <int>, watchers <int>,
## # default_branch <chr>, homepage <chr>
ASOIAF characters
Let’s look at got_chars, which is a list of information on the point-of-view characters from the first five books in A Song of Ice and Fire by George R.R. Martin.
Each element corresponds to one character and contains 18 sub-elements which are named atomic vectors of various lengths and types. We start in the same way, first by creating a data frame and then by unnesting each component into a column:
chars <- tibble(char = got_chars)
chars
## # A tibble: 30 x 1
## char
## <list>
## 1 <named list [18]>
## 2 <named list [18]>
## 3 <named list [18]>
## 4 <named list [18]>
## 5 <named list [18]>
## 6 <named list [18]>
## 7 <named list [18]>
## 8 <named list [18]>
## 9 <named list [18]>
## 10 <named list [18]>
## # … with 20 more rows
chars2 <- chars %>%
unnest_wider(char)
chars2
## # A tibble: 30 x 18
## url id name gender culture born died alive titles aliases father
## <chr> <int> <chr> <chr> <chr> <chr> <chr> <lgl> <list> <list> <chr>
## 1 http… 1022 Theo… Male "Ironb… "In … "" TRUE <chr … <chr [… ""
## 2 http… 1052 Tyri… Male "" "In … "" TRUE <chr … <chr [… ""
## 3 http… 1074 Vict… Male "Ironb… "In … "" TRUE <chr … <chr [… ""
## 4 http… 1109 Will Male "" "" "In … FALSE <chr … <chr [… ""
## 5 http… 1166 Areo… Male "Norvo… "In … "" TRUE <chr … <chr [… ""
## 6 http… 1267 Chett Male "" "At … "In … FALSE <chr … <chr [… ""
## 7 http… 1295 Cres… Male "" "In … "In … FALSE <chr … <chr [… ""
## 8 http… 130 Aria… Female "Dorni… "In … "" TRUE <chr … <chr [… ""
## 9 http… 1303 Daen… Female "Valyr… "In … "" TRUE <chr … <chr [… ""
## 10 http… 1319 Davo… Male "Weste… "In … "" TRUE <chr … <chr [… ""
## # … with 20 more rows, and 7 more variables: mother <chr>, spouse <chr>,
## # allegiances <list>, books <list>, povBooks <list>, tvSeries <list>,
## # playedBy <list>
This is more complex than gh_users because some component of char are themselves a list, giving us a collection of list-columns:
chars2 %>%
select_if(is.list)
## # A tibble: 30 x 7
## titles aliases allegiances books povBooks tvSeries playedBy
## <list> <list> <list> <list> <list> <list> <list>
## 1 <chr [3]> <chr [4]> <chr [1]> <chr [3]> <chr [2]> <chr [6]> <chr [1]>
## 2 <chr [2]> <chr [11]> <chr [1]> <chr [2]> <chr [4]> <chr [6]> <chr [1]>
## 3 <chr [2]> <chr [1]> <chr [1]> <chr [3]> <chr [2]> <chr [1]> <chr [1]>
## 4 <chr [1]> <chr [1]> <NULL> <chr [1]> <chr [1]> <chr [1]> <chr [1]>
## 5 <chr [1]> <chr [1]> <chr [1]> <chr [3]> <chr [2]> <chr [2]> <chr [1]>
## 6 <chr [1]> <chr [1]> <NULL> <chr [2]> <chr [1]> <chr [1]> <chr [1]>
## 7 <chr [1]> <chr [1]> <NULL> <chr [2]> <chr [1]> <chr [1]> <chr [1]>
## 8 <chr [1]> <chr [1]> <chr [1]> <chr [4]> <chr [1]> <chr [1]> <chr [1]>
## 9 <chr [5]> <chr [11]> <chr [1]> <chr [1]> <chr [4]> <chr [6]> <chr [1]>
## 10 <chr [4]> <chr [5]> <chr [2]> <chr [1]> <chr [3]> <chr [5]> <chr [1]>
## # … with 20 more rows
What you do next will depend on the purposes of the analysis. Maybe you want a row for every book and TV series that the character appears in:
chars2 %>%
select(name, books, tvSeries) %>%
pivot_longer(c(books, tvSeries), names_to = "media", values_to = "value") %>%
unnest_longer(value)
## # A tibble: 180 x 3
## name media value
## <chr> <chr> <chr>
## 1 Theon Greyjoy books A Game of Thrones
## 2 Theon Greyjoy books A Storm of Swords
## 3 Theon Greyjoy books A Feast for Crows
## 4 Theon Greyjoy tvSeries Season 1
## 5 Theon Greyjoy tvSeries Season 2
## 6 Theon Greyjoy tvSeries Season 3
## 7 Theon Greyjoy tvSeries Season 4
## 8 Theon Greyjoy tvSeries Season 5
## 9 Theon Greyjoy tvSeries Season 6
## 10 Tyrion Lannister books A Feast for Crows
## # … with 170 more rows
Or maybe you want to build a table that lets you match title to name:
chars2 %>%
select(name, title = titles) %>%
unnest_longer(title)
## # A tibble: 60 x 2
## name title
## <chr> <chr>
## 1 Theon Greyjoy "Prince of Winterfell"
## 2 Theon Greyjoy "Captain of Sea Bitch"
## 3 Theon Greyjoy "Lord of the Iron Islands (by law of the green lands)"
## 4 Tyrion Lannister "Acting Hand of the King (former)"
## 5 Tyrion Lannister "Master of Coin (former)"
## 6 Victarion Greyjoy "Lord Captain of the Iron Fleet"
## 7 Victarion Greyjoy "Master of the Iron Victory"
## 8 Will ""
## 9 Areo Hotah "Captain of the Guard at Sunspear"
## 10 Chett ""
## # … with 50 more rows
Again, we could rewrite using unnest_auto(). This is convenient for exploration, but I wouldn’t rely on it in the long term - unnest_auto() has the undesirable property that it will always succeed. That means if your data structure changes, unnest_auto() will continue to work, but might give very different output that causes cryptic failures from downstream functions.
tibble(char = got_chars) %>%
unnest_auto(char) %>%
select(name, title = titles) %>%
unnest_auto(title)
## Using `unnest_wider(char)`; elements have 18 names in common
## Using `unnest_longer(title)`; no element has names
## # A tibble: 60 x 2
## name title
## <chr> <chr>
## 1 Theon Greyjoy "Prince of Winterfell"
## 2 Theon Greyjoy "Captain of Sea Bitch"
## 3 Theon Greyjoy "Lord of the Iron Islands (by law of the green lands)"
## 4 Tyrion Lannister "Acting Hand of the King (former)"
## 5 Tyrion Lannister "Master of Coin (former)"
## 6 Victarion Greyjoy "Lord Captain of the Iron Fleet"
## 7 Victarion Greyjoy "Master of the Iron Victory"
## 8 Will ""
## 9 Areo Hotah "Captain of the Guard at Sunspear"
## 10 Chett ""
## # … with 50 more rows
May the force be with you
sw_people, sw_films, sw_species, sw_planets, sw_starships and sw_vehicles are interrelated lists in the repurrrsive package about entities in the Star Wars Universe retrieved from the Star Wars API using the package rwars.
map_chr(sw_films, "title")
## [1] "A New Hope" "Attack of the Clones"
## [3] "The Phantom Menace" "Revenge of the Sith"
## [5] "Return of the Jedi" "The Empire Strikes Back"
## [7] "The Force Awakens"
Use your knowledge of rectangling with tidyr to extract relevant data of interest from these data frames to complete the following exercises.
Generate a visualization of the distribution of average height for each species in the Star Wars universe.
Click for the solution
sw_speciescontains one element for each species in the database, so we should useunnest_wider()orhoist()to extract the required elements.# clean up sw_species so it is one-row-per-species sw_height <- tibble(sw_species) %>% hoist(sw_species, height = "average_height" ) %>% # fix height to be a numeric column mutate(height = parse_number(height))## Warning: 3 parsing failures. ## row col expected actual ## 19 -- a number unknown ## 29 -- a number unknown ## 35 -- a number n/asw_height## # A tibble: 37 x 2 ## height sw_species ## <dbl> <list> ## 1 300 <named list [14]> ## 2 66 <named list [14]> ## 3 200 <named list [14]> ## 4 160 <named list [14]> ## 5 100 <named list [14]> ## 6 180 <named list [14]> ## 7 180 <named list [14]> ## 8 190 <named list [14]> ## 9 120 <named list [14]> ## 10 100 <named list [14]> ## # … with 27 more rows# generate a histogram ggplot(data = sw_height, mapping = aes(x = height)) + geom_histogram() + labs( x = "Height (in centimeters)", y = "Number of species" )## `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.## Warning: Removed 3 rows containing non-finite values (stat_bin).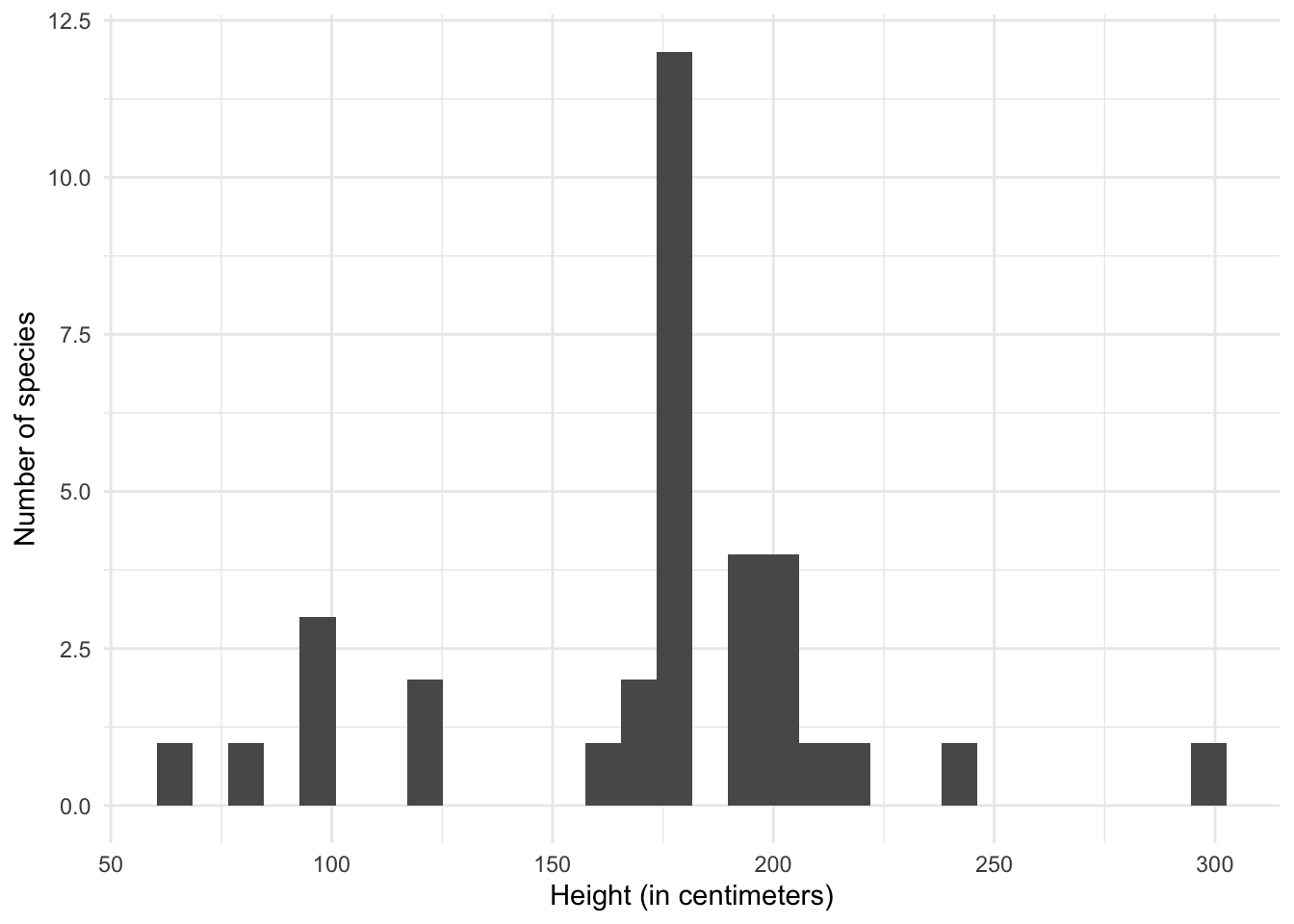
Generate a bar chart showing the number of film appearances made by each character in
sw_peoplewho made at least three film appearances.Click for the solution
Each element of
sw_peoplecontains one character. Thefilmselement within each character is a character vector containing one value for each film in which the character appeared. This required two separateunnest_*()operations to get the data in the proper form.# unnest the data sw_people_df <- tibble(sw_people) %>% unnest_wider(sw_people) %>% unnest_longer(films) sw_people_df## # A tibble: 173 x 16 ## name height mass hair_color skin_color eye_color birth_year gender ## <chr> <chr> <chr> <chr> <chr> <chr> <chr> <chr> ## 1 Luke Skywalker 172 77 blond fair blue 19BBY male ## 2 Luke Skywalker 172 77 blond fair blue 19BBY male ## 3 Luke Skywalker 172 77 blond fair blue 19BBY male ## 4 Luke Skywalker 172 77 blond fair blue 19BBY male ## 5 Luke Skywalker 172 77 blond fair blue 19BBY male ## 6 C-3PO 167 75 n/a gold yellow 112BBY n/a ## 7 C-3PO 167 75 n/a gold yellow 112BBY n/a ## 8 C-3PO 167 75 n/a gold yellow 112BBY n/a ## 9 C-3PO 167 75 n/a gold yellow 112BBY n/a ## 10 C-3PO 167 75 n/a gold yellow 112BBY n/a ## # … with 163 more rows, and 8 more variables: homeworld <chr>, films <chr>, ## # species <chr>, vehicles <list>, starships <list>, created <chr>, ## # edited <chr>, url <chr># summarize the data frame and graph the bar chart sw_people_df %>% count(name) %>% filter(n >= 3) %>% ggplot(mapping = aes(x = fct_reorder(.f = name, .x = n), y = n)) + geom_col() + coord_flip() + labs( title = "Number of appearances in the Star Wars cinematic universe", subtitle = "As of December 31, 2015", x = NULL, y = "Number of film appearances" )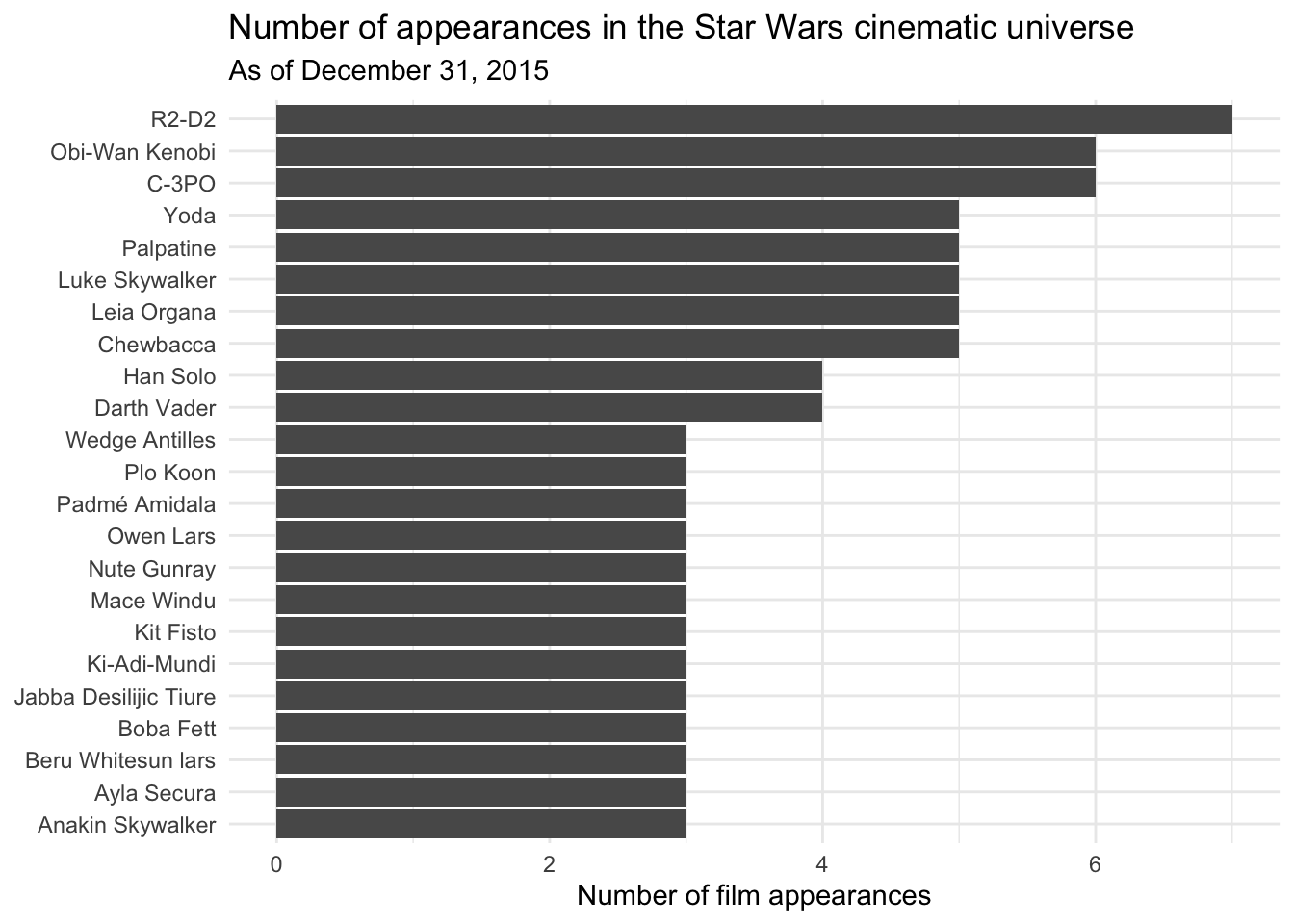
Acknowledgments
- Examples and data files drawn from Jenny Bryan’s
purrrtutorial - Examples and data files also drawn from the rectangling vignette in
tidyr.
Session Info
devtools::session_info()
## ─ Session info ───────────────────────────────────────────────────────────────
## setting value
## version R version 4.1.0 (2021-05-18)
## os macOS Big Sur 10.16
## system x86_64, darwin17.0
## ui X11
## language (EN)
## collate en_US.UTF-8
## ctype en_US.UTF-8
## tz America/Chicago
## date 2021-11-15
##
## ─ Packages ───────────────────────────────────────────────────────────────────
## package * version date lib source
## assertthat 0.2.1 2019-03-21 [1] CRAN (R 4.1.0)
## backports 1.2.1 2020-12-09 [1] CRAN (R 4.1.0)
## blogdown 1.4 2021-07-23 [1] CRAN (R 4.1.0)
## bookdown 0.23 2021-08-13 [1] CRAN (R 4.1.0)
## broom 0.7.9 2021-07-27 [1] CRAN (R 4.1.0)
## bslib 0.3.1 2021-10-06 [1] CRAN (R 4.1.0)
## cachem 1.0.6 2021-08-19 [1] CRAN (R 4.1.0)
## callr 3.7.0 2021-04-20 [1] CRAN (R 4.1.0)
## cellranger 1.1.0 2016-07-27 [1] CRAN (R 4.1.0)
## cli 3.1.0 2021-10-27 [1] CRAN (R 4.1.0)
## colorspace 2.0-2 2021-06-24 [1] CRAN (R 4.1.0)
## crayon 1.4.2 2021-10-29 [1] CRAN (R 4.1.0)
## DBI 1.1.1 2021-01-15 [1] CRAN (R 4.1.0)
## dbplyr 2.1.1 2021-04-06 [1] CRAN (R 4.1.0)
## desc 1.3.0 2021-03-05 [1] CRAN (R 4.1.0)
## devtools 2.4.2 2021-06-07 [1] CRAN (R 4.1.0)
## digest 0.6.28 2021-09-23 [1] CRAN (R 4.1.0)
## dplyr * 1.0.7 2021-06-18 [1] CRAN (R 4.1.0)
## ellipsis 0.3.2 2021-04-29 [1] CRAN (R 4.1.0)
## evaluate 0.14 2019-05-28 [1] CRAN (R 4.1.0)
## fansi 0.5.0 2021-05-25 [1] CRAN (R 4.1.0)
## fastmap 1.1.0 2021-01-25 [1] CRAN (R 4.1.0)
## forcats * 0.5.1 2021-01-27 [1] CRAN (R 4.1.0)
## fs 1.5.0 2020-07-31 [1] CRAN (R 4.1.0)
## generics 0.1.1 2021-10-25 [1] CRAN (R 4.1.0)
## ggplot2 * 3.3.5 2021-06-25 [1] CRAN (R 4.1.0)
## glue 1.5.0 2021-11-07 [1] CRAN (R 4.1.0)
## gtable 0.3.0 2019-03-25 [1] CRAN (R 4.1.0)
## haven 2.4.3 2021-08-04 [1] CRAN (R 4.1.0)
## here 1.0.1 2020-12-13 [1] CRAN (R 4.1.0)
## hms 1.1.1 2021-09-26 [1] CRAN (R 4.1.0)
## htmltools 0.5.2 2021-08-25 [1] CRAN (R 4.1.0)
## httr * 1.4.2 2020-07-20 [1] CRAN (R 4.1.0)
## jquerylib 0.1.4 2021-04-26 [1] CRAN (R 4.1.0)
## jsonlite 1.7.2 2020-12-09 [1] CRAN (R 4.1.0)
## knitr 1.33 2021-04-24 [1] CRAN (R 4.1.0)
## lifecycle 1.0.1 2021-09-24 [1] CRAN (R 4.1.0)
## lubridate 1.7.10 2021-02-26 [1] CRAN (R 4.1.0)
## magrittr 2.0.1 2020-11-17 [1] CRAN (R 4.1.0)
## memoise 2.0.0 2021-01-26 [1] CRAN (R 4.1.0)
## modelr 0.1.8 2020-05-19 [1] CRAN (R 4.1.0)
## munsell 0.5.0 2018-06-12 [1] CRAN (R 4.1.0)
## pillar 1.6.4 2021-10-18 [1] CRAN (R 4.1.0)
## pkgbuild 1.2.0 2020-12-15 [1] CRAN (R 4.1.0)
## pkgconfig 2.0.3 2019-09-22 [1] CRAN (R 4.1.0)
## pkgload 1.2.1 2021-04-06 [1] CRAN (R 4.1.0)
## prettyunits 1.1.1 2020-01-24 [1] CRAN (R 4.1.0)
## processx 3.5.2 2021-04-30 [1] CRAN (R 4.1.0)
## ps 1.6.0 2021-02-28 [1] CRAN (R 4.1.0)
## purrr * 0.3.4 2020-04-17 [1] CRAN (R 4.1.0)
## R6 2.5.1 2021-08-19 [1] CRAN (R 4.1.0)
## Rcpp 1.0.7 2021-07-07 [1] CRAN (R 4.1.0)
## readr * 2.0.2 2021-09-27 [1] CRAN (R 4.1.0)
## readxl 1.3.1 2019-03-13 [1] CRAN (R 4.1.0)
## remotes 2.4.0 2021-06-02 [1] CRAN (R 4.1.0)
## reprex 2.0.1 2021-08-05 [1] CRAN (R 4.1.0)
## repurrrsive * 1.0.0 2019-07-15 [1] CRAN (R 4.1.0)
## rlang 0.4.12 2021-10-18 [1] CRAN (R 4.1.0)
## rmarkdown 2.11 2021-09-14 [1] CRAN (R 4.1.0)
## rprojroot 2.0.2 2020-11-15 [1] CRAN (R 4.1.0)
## rstudioapi 0.13 2020-11-12 [1] CRAN (R 4.1.0)
## rvest 1.0.1 2021-07-26 [1] CRAN (R 4.1.0)
## sass 0.4.0 2021-05-12 [1] CRAN (R 4.1.0)
## scales 1.1.1 2020-05-11 [1] CRAN (R 4.1.0)
## sessioninfo 1.1.1 2018-11-05 [1] CRAN (R 4.1.0)
## stringi 1.7.5 2021-10-04 [1] CRAN (R 4.1.0)
## stringr * 1.4.0 2019-02-10 [1] CRAN (R 4.1.0)
## testthat 3.0.4 2021-07-01 [1] CRAN (R 4.1.0)
## tibble * 3.1.6 2021-11-07 [1] CRAN (R 4.1.0)
## tidyr * 1.1.4 2021-09-27 [1] CRAN (R 4.1.0)
## tidyselect 1.1.1 2021-04-30 [1] CRAN (R 4.1.0)
## tidyverse * 1.3.1 2021-04-15 [1] CRAN (R 4.1.0)
## tzdb 0.1.2 2021-07-20 [1] CRAN (R 4.1.0)
## usethis 2.0.1 2021-02-10 [1] CRAN (R 4.1.0)
## utf8 1.2.2 2021-07-24 [1] CRAN (R 4.1.0)
## vctrs 0.3.8 2021-04-29 [1] CRAN (R 4.1.0)
## withr 2.4.2 2021-04-18 [1] CRAN (R 4.1.0)
## xfun 0.25 2021-08-06 [1] CRAN (R 4.1.0)
## xml2 1.3.2 2020-04-23 [1] CRAN (R 4.1.0)
## yaml 2.2.1 2020-02-01 [1] CRAN (R 4.1.0)
##
## [1] /Library/Frameworks/R.framework/Versions/4.1/Resources/library